Part 1 explored the innovations that are reshaping the peer review process, including community and third-party services that are expanding the reviewer pool, as well as preprint servers, overlay journals, and postpublication forums that serve as examples of a more open and transparent ecosystem. Part 2 highlights the technology initiatives that streamline processes and enable experimentation in peer review. These technologies, including communication protocols, messaging services, embedded XML, and persistent identifiers, provide the necessary digital infrastructure. Also included are four key use cases that illustrate how decentralized peer review is an effective and innovative contributor to the scholarly publishing ecosystem. Each use case highlights the potential benefits of these new models, including increased transparency, faster turnaround times, and greater inclusivity. By integrating community-driven platforms, open protocols, and diversity-focused initiatives, these use cases provide a blueprint for the future of peer review.
Technology and System-to-System Communication Protocols in Peer Review: Enhancing Efficiency and Transparency
The digital era has reshaped scholarly publishing, and system-to-system communication protocols have become integral to modern peer review workflows. These protocols enable seamless data exchange between research platforms, preprint servers, journals, and peer review services, allowing reviews and research metadata to follow manuscripts across various stages of publication. This ensures peer review processes are portable, efficient, and accessible. Key technologies facilitating these exchanges include Manuscript Exchange Common Approach (MECA), Confederation of Open Access Repositories (COAR) Notify, and DocMaps, all of which promote interoperability in scholarly communication.
MECA
MECA, a NISO Recommended Practice, streamlines the transfer of research manuscripts between systems. MECA is a standardized protocol that defines how to package and transfer manuscript files and associated metadata from one system to another. Whether a manuscript is being moved between preprint servers and journals, or across different submission systems, MECA ensures that all relevant data travels with the manuscript, reducing redundancies and the need for re-entering information.
One of MECA’s primary uses is to facilitate cascading workflows, where manuscripts move seamlessly between different stages of submission and review. MECA supports 3 primary use cases:
- Submission System to Submission System. This enables cross-publisher transfers, allowing manuscripts to move easily from one journal to another, while maintaining the full peer review history.
- Preprint System to/from Submission System. MECA supports the transfer of manuscripts from preprint servers to formal submission systems, addressing the growing popularity of pre-review distribution on platforms like bioRxiv and arXiv.
- Authoring System to Submission System. By simplifying the connection between authoring platforms and journals, MECA helps authors quickly submit their work to the preprint server or journal of their choice.
MECA is built around the key principles of minimizing repeated data entry and maintaining interoperability. It promotes consistency and reliability in manuscript exchanges, eliminating redundant efforts and making the entire submission and review process faster and more convenient. For more details about MECA, visit the official NISO MECA webpage.1
COAR Notify: Connecting Repositories to External Services
COAR Notify, developed by COAR, is an initiative that supports the seamless integration of research outputs hosted in repositories and preprint servers with external services, such as overlay journals and peer review platforms. COAR Notify provides a decentralized, interoperable system that allows research outputs like preprints to be linked with peer review services and journals, ensuring that data travels efficiently between different platforms.
The primary aim of COAR Notify is to reduce the technological barriers between systems, enabling repositories and review services to participate in the evolving publish, review, curate model for scholarly communication. Some key use cases include:
- Allowing authors to request peer reviews directly from a repository when they deposit a preprint.
- Enabling authors to request publication by an overlay journal that sits on top of a repository.
- Linking datasets from one repository to articles housed in another, promoting more cohesive research networks.
COAR Notify’s interoperability is essential for developing community-led peer review platforms. Like Peer Community In (PCI) and PREreview, allowing them to scale and engage with the broader publishing ecosystem. By putting these connections in place, COAR Notify helps bridge the gap between repositories and review services, making peer reviewed preprints more accessible. You can learn more about COAR Notify here.2
DocMaps: A Breadcrumb Trail for Research Evaluation
Developed by MIT’s Knowledge Futures Group, DocMaps offers a framework for creating machine-readable documentation of the editorial and peer review processes that research manuscripts undergo. DocMaps provides a structured way to capture the editorial journey of a manuscript, including key details such as peer reviews, editorial decisions, and revisions. This metadata can then be embedded into the document, allowing other systems to interpret and use it.
The concept behind DocMaps is akin to leaving a breadcrumb trail for research, documenting every step of the peer review and editorial processes. The system can record details like:
- When and where a manuscript was submitted
- What quality check tools were administered
- Which reviewers were involved and their feedback
- Revisions requested and subsequent responses from the authors
DocMaps ensures this information is machine-readable, meaning that other platforms—such as indexing services, repositories, or funders—can extract the data to analyze the quality and transparency of the peer review process. By documenting the editorial path of research in a standardized format, DocMaps promotes greater transparency and accountability in scholarly publishing. This benefits not only readers but also funders and institutions looking to assess the rigor of the peer review process. For more information on DocMaps, visit DocMaps Knowledge Futures.3
Persistent Identifiers: Building Trust and Integrity in Research
Alongside these system-to-system protocols, the use of persistent identifiers (PIDs) is crucial for ensuring interoperability as well as the trustworthiness and integrity of research workflows. PIDs are standardized unique identifiers assigned to individuals, institutions, and research outputs, allowing them to be tracked and referenced across different platforms. In the context of peer review, PIDs are essential for ensuring that data can be accurately linked and verified, ensuring trust among authors, reviewers, and readers.
Here are some of the most important PIDs used in scholarly publishing:
- ORCID. ORCID (Open Researcher and Contributor ID) provides a unique, persistent identifier for individual researchers, allowing their work to be easily linked across platforms. ORCID helps to disambiguate authors with similar names and ensures that contributions to research—whether authorship, review, or editing—are correctly attributed. ORCID is particularly important in peer review, where it can be used to verify the identities of reviewers and ensure the integrity of the review process. For more information on ORCID, visit ORCID.org.
- ROR. ROR (Research Organization Registry) is a persistent identifier for research organizations, including funding organizations, ensuring that institutional affiliations are correctly attributed in research outputs. ROR helps to track the contributions of institutions to research and ensures that organizational data remains consistent, even in cases of name changes or mergers. It allows research outputs to be accurately connected to the institutions that supported them. You can explore more about ROR at ROR.org.
- DOIs. DOIs (Digital Object Identifiers) are assigned to research outputs such as articles, datasets, and even peer reviews. DOIs provide a permanent link to a research object, making it easy to find, cite, and reuse. In peer review, DOIs can be assigned to individual reviews, ensuring that reviewers receive proper credit for their contributions. Two of the most prominent organizations that manage DOIs are CrossRef and DataCite. Crossref provides DOIs for scholarly content, enabling accurate citation, linking, and metadata sharing to support research discovery and interoperability across scholarly publishing platforms. DataCite plays a critical role in assigning DOIs to datasets, helping to promote open data and reproducibility in research. More information on DOIs can be found at CrossRef4 and DataCite.5
- RRIDs. RRIDs (Research Resource Identifiers) are persistent identifiers for biological resources, such as cell lines, antibodies, or model organisms, used in scientific experiments. By using RRIDs, researchers can ensure that these resources are consistently referenced across studies, improving the reproducibility of scientific findings. RRIDs provide transparency in reporting experimental methods and materials, helping future researchers replicate the experiments more accurately. Learn more about RRIDs at RRIDs.org.
Benefits of System-to-System Protocols and PIDs
The integration of system-to-system communication protocols and persistent identifiers offers numerous benefits for the scholarly publishing ecosystem. Most importantly, it helps make research workflows more efficient, transparent, and trustworthy, creating a collaborative and open research environment. Protocols such as MECA, COAR Notify, and DocMaps enable seamless exchange of metadata and peer review information across systems, allowing manuscripts to flow smoothly between preprint servers, submission systems, and journals. At the same time, protocols like DocMaps promote transparency by generating a machine-readable record of each step in a manuscript’s editorial journey, allowing readers, funders, and institutions to understand the review process and fostering trust in peer review.
Persistent identifiers are integral to this ecosystem, as they enable precise identification and connection of authors, institutions, and research outputs across different platforms. By embedding PIDs within system-to-system communications, interoperability is enhanced, ensuring that contributions are properly tracked and attributed throughout the publishing lifecycle. This not only facilitates efficient collaboration between preprint servers, journals, and peer review services but also establishes accountability and reinforces the integrity of research outputs. Together, these protocols and PIDs provide the foundation for a robust, transparent, and efficient research infrastructure.
Mapping the Preprint Metadata Transfer Ecosystem: Understanding the Complexity of Decentralized Peer Review
As these various services and systems have evolved—independent peer review, preprint servers, overlay journals, communications protocols, and PIDs—there is a need to better understand how all of this can fit together. The Map the Preprint Metadata Transfer Ecosystem6 project seeks to address this need by diagramming how peer reviews are conducted, transferred, and integrated across multiple systems. This project, co-organized by Europe PMC and ASAPbio, brings together stakeholders from preprint review projects, infrastructure providers, publishers, and funders to determine the key elements of preprint review metadata and the mechanisms for sharing this information across platforms.
The initiative provides a comprehensive roadmap of how reviews and metadata flow between preprint servers, overlay journals, and other peer review services, illustrating the complex interactions required to maintain a transparent, interoperable peer review ecosystem. By mapping out these processes, the project aims to create a shared understanding of the pathways used to transfer peer review metadata and identify the challenges and gaps that need to be addressed to ensure interoperability across the various systems involved in scholarly communication. This roadmap shows how different protocols and systems interact to create a decentralized peer review ecosystem.
Metadata Elements and Transfer Pathways
The metadata transfer ecosystem encompasses a wide range of elements, from the basic metadata that accompanies a manuscript (e.g., title, authors, submission date) to more complex elements like peer review reports, reviewer identities, and editorial decisions. One of the key challenges the project identifies is the lack of standardization across platforms. Each system has its own approach to handling metadata, which can lead to inconsistencies and difficulties in transferring data between platforms. Central to this effort are system-to-system communication protocols (MECA, COAR Notify, DocMaps) used to transfer metadata and files.
As manuscripts and their associated reviews move through the publishing ecosystem—from preprint servers to overlay journals and beyond—metadata needs to be transferred along with the content to ensure that metadata, including reviews, are available to subsequent platforms and stakeholders. The project is working to harmonize metadata standards across systems to ensure that peer review data can be easily shared and interpreted. For example, the use of PIDs, such as DOIs, ORCID IDs, and RRIDs, is critical for maintaining the integrity of metadata transfers. PIDs ensure that the correct individuals, institutions, and research objects are consistently identified, regardless of which system the metadata passes through. These identifiers are essential for tracking peer reviews across platforms and ensuring that all contributions are properly attributed.
An Example Workflow
The project’s metadata transfer road mapping process involves creating clear workflows that show how services, repositories, and protocols interact to share information. This road mapping helps to clarify how metadata can flow smoothly between different platforms, enabling more efficient peer review processes. One typical workflow in the metadata transfer ecosystem might look like this:
- A researcher deposits a preprint in a repository such as bioRxiv or arXiv. They include ORCIDs for all contributors and RORs in the affiliations.
- The preprint is assigned a Crossref DOI, making it citable and trackable across platforms.
- The author requests a peer review through an external peer review service such as PREreview or Sciety.
- Once the review is completed, it is linked back to the preprint using COAR Notify, which ensures that the review metadata is available to any platform accessing the preprint. The reviewer is given recognition for their review via their ORCID.
- The review process is documented using DocMaps, which creates a machine-readable record of the peer review and editorial decisions.
- The preprint, along with its reviews, might be transferred to a journal using MECA, allowing the manuscript to be published in a traditional journal, while retaining all its metadata.
- The peer review reports and metadata are made openly accessible via repositories such as Zenodo or aggregators like preLights, ensuring that readers, funders, and other researchers can view the complete history of the manuscript’s evaluation.
This workflow demonstrates how the various components of the metadata transfer ecosystem work together to facilitate open, transparent peer review (Figure). By ensuring that peer reviews and metadata are transferred efficiently between systems, this ecosystem enables a decentralized model of peer review, where multiple platforms can collaborate to evaluate research without sacrificing transparency or accountability.
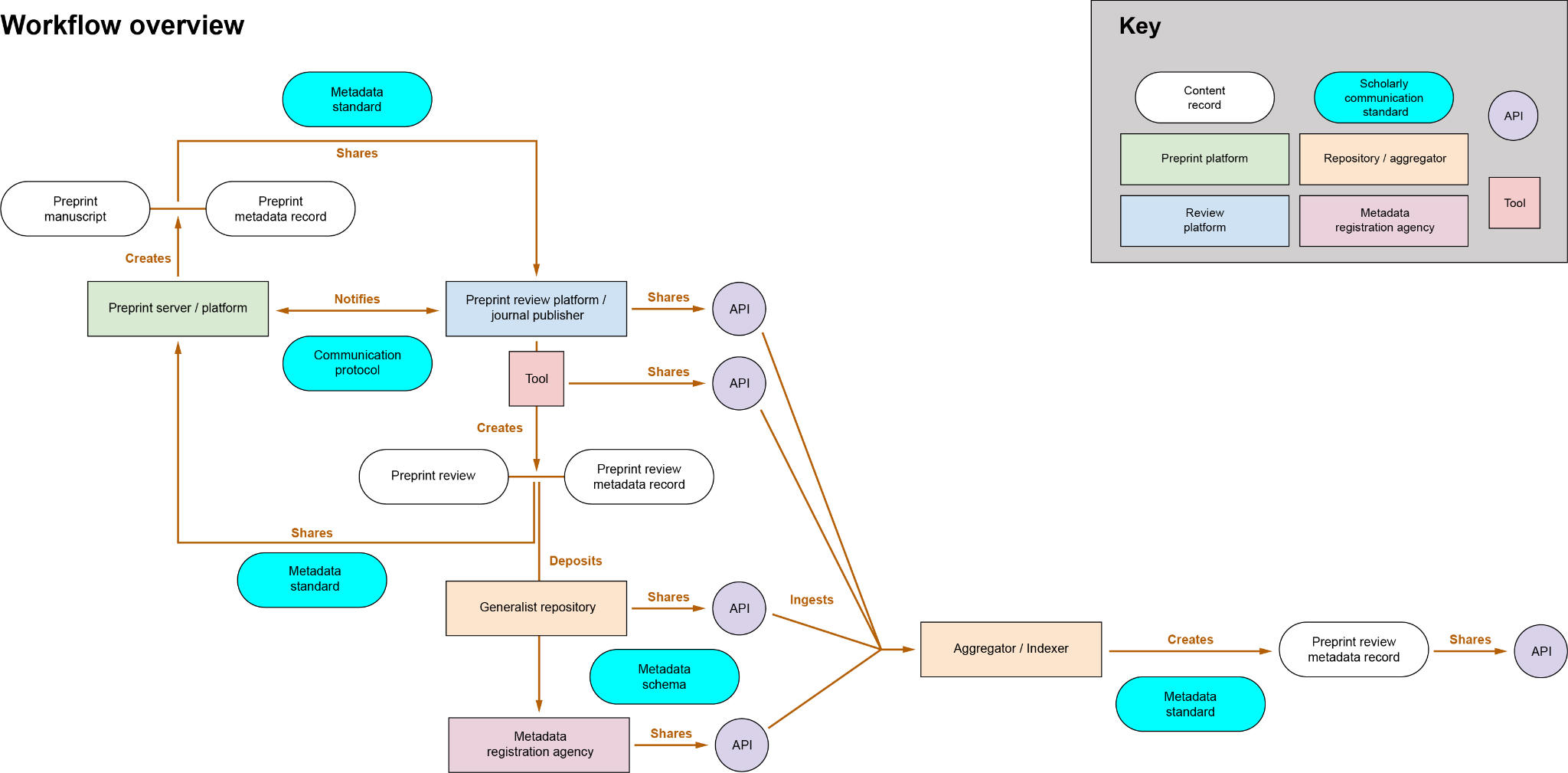
For more information about the Map the Preprint Metadata Transfer Ecosystem, details on the project can be found here.7
Use Case 1: Community Peer Review in Traditional Workflows
Collaboration between PREreview and the journal Current Research in Neurobiology (CRNEUR) exemplifies how community-based peer review can be integrated into the traditional journal workflow, adding a layer of transparency and inclusivity to the review process. As mentioned, PREreview is an open platform that allows researchers to review preprints and engage in collaborative discussions about scientific manuscripts. In this initiative with CRNEUR, community-based reviews are conducted alongside traditional peer reviews, providing authors with more comprehensive and diverse feedback on their work.
The PREreview and CRNEUR pilot project is a particularly exciting example of how community peer review can be used to complement traditional journal peer review. When a manuscript is submitted to CRNEUR, it is also posted on a preprint server, where members of the PREreview community can participate in a PREreview Live Review session. These live sessions involve 2 PREreview team members who lead a discussion with participants, collaboratively taking notes and offering constructive feedback on the manuscript. The feedback is then compiled into a review, which is shared with the authors and published on the PREreview platform under an open license.
This collaborative review process has several benefits. First, it allows for a more diverse range of perspectives, as researchers from different backgrounds and career stages can participate in the review. Second, it increases the transparency of the review process, as the feedback is made publicly available. Finally, it provides authors with more detailed and actionable feedback, which they can use to improve their work before submitting it for final publication. The CRNEUR editorial team considers these community reviews alongside the traditional peer reviews, allowing for a more holistic evaluation of the manuscript.
More information on the PREreview model and its impact on peer review can be found here.8
Use Case 2: Preprint Server Review Using Open Protocols
Preprints allow researchers to share their findings with the scientific community and the public much earlier than in traditional publishing models. bioRxiv, one of the leading preprint servers, has pioneered the use of open protocols to facilitate decentralized peer review, making it easier for researchers to access and engage with preprint evaluations.
One of the key innovations introduced by bioRxiv is its review dashboard, which aggregates feedback from various sources, including journal-organized peer reviews, community discussions, and social media mentions. This dashboard provides a comprehensive view of how a preprint is being received by the scientific community, allowing readers to quickly assess the credibility and relevance of the research. The reviews and comments are linked to the preprint abstract, making it easy for users to access the feedback and join the conversation.
bioRxiv’s use of open protocols, such as MECA and DocMaps, enable the smooth transfer of reviews and comments across platforms. These protocols ensure the metadata associated with a manuscript, including its review history, is easily accessible and transferable between different systems. This interoperability is essential for creating a seamless peer review ecosystem, where feedback can follow a manuscript as it moves through different stages of the publication process.
Additionally, bioRxiv has integrated Hypothesis, a web-based annotation tool that allows users to leave comments directly on preprints. This feature promotes active engagement with the research, as readers can provide feedback in real-time, highlighting strengths, identifying potential flaws, and suggesting improvements. By making the review process more interactive and transparent, bioRxiv is helping to accelerate the dissemination of knowledge and foster a more collaborative approach to scientific discovery.
bioRxiv’s use of open protocols and review aggregation tools represents a step forward in making peer review more transparent and accessible. By providing a platform for decentralized feedback, bioRxiv is helping to break down the barriers between researchers and reviewers, creating a more open and inclusive scholarly publishing ecosystem. Learn more about bioRxiv’s efforts here.9
Use Case 3: Aggregation of Preprint Reviews from Disparate Sources
It can be difficult to find related research when it is scattered across various preprint servers and repositories. Two initiatives, Early Evidence Base (EEB) by EMBO, and Sciety by eLife are platforms designed to aggregate peer reviewed preprints, making them accessible, citable, and easily discoverable.
EEB empowers researchers, especially early-career scientists, by enabling them to showcase their scientific contributions even before formal journal publication. By providing a centralized repository for peer reviewed preprints, EEB not only increases the visibility of these works but also facilitates their citation and use in ongoing research.
Researchers can cite reviewed preprints by appending the preprint DOI to the URL (e.g., https://eeb.embo.org/doi/). For example, a reviewed preprint might be cited as follows:
Moussa AT, Cosenza MR, Wohlfromm T, Brobeil K, Hill A, Patrizi A, Müller-Decker K, Holland-Letz T, Jauch A, Kraft B, Krämer A. (2023). STIL overexpression shortens lifespan and reduces tumor formation in mice. bioRxiv doi.org/10.1101/2023.12.04.569842, peer reviewed by Review Commons eeb.embo.org/doi/10.1101/2023.12.04.569842.
Beyond just aggregating preprints, EEB is also a technological experiment aimed at advancing transparency and accessibility in the scholarly publishing process. It leverages the DocMaps format to send machine-readable information about the editorial process, enabling different stakeholders, such as readers, funders, and institutions, to visualize the peer review history of a manuscript in a standardized way. This visualization allows readers to access feedback and author replies in a clear, user-friendly format, enhancing understanding of the review process and the development of research findings.
EEB serves as an experimental platform for decentralized peer review. It aggregates, publishes, exchanges, filters, and mines “reviewed preprints” across a distributed network, thereby expanding the conventional publishing model to include these innovative publishing objects. Through its combination of system-to-system communication protocols and persistent identifiers, EEB exemplifies a decentralized approach to scholarly publishing. More details on the EEB platform can be found here.10
Another platform, Sciety, developed by eLife, aggregates peer reviews and public evaluations of preprints from trusted groups.11 Like EEB, Sciety helps researchers navigate the growing volume of preprints by bringing together evaluations from a variety of sources, such as ReviewCommons, PREreview, ASAPbio’s Crowd Review, and eLife’s own peer review efforts. Sciety provides a comprehensive view of a preprint’s credibility by presenting associated evaluations in context alongside a preprint, helping researchers identify the most relevant and impactful studies.
Sciety promotes open and transparent evaluations, ensuring that all reviews are accessible to the public. Researchers can view multiple community evaluations of the same preprint, offering a diversity of perspectives not possible in the traditional publishing process, and making it easier to assess the quality and significance of the research. Scientists can create and share themed lists/collections of preprints they find interesting, helping to increase the visibility of important research and shape the preprint landscape.
Sciety’s aggregation and display of preprint peer reviews provide a resource for researchers looking to understand the broader impact of a preprint, often before it is published in a journal. Through public evaluation, Sciety helps build trust in the peer review process. For more details on Sciety’s platform, visit Sciety.org.
Use Case 4: Bringing Diversity to Peer Review
Diversity, equity, and inclusion (DEI) initiatives are increasingly being recognized as essential for improving the peer review process. As discussed, many organizations are actively working to train and support underrepresented groups in academia, ensuring that a more diverse range of voices is included in the evaluation of research. Following are 2 examples of these efforts.
The collaboration between PREreview and the Howard Hughes Medical Institute (HHMI) centers on creating a more diverse, transparent and accountable peer review process through targeted training initiatives. HHMI, a leading biomedical research organization, supports an inclusive scientific community and has partnered with PREreview to pilot a Transparent and Accountable Peer Review Training Program. This initiative, aimed at graduate students and postdoctoral researchers within HHMI labs, focuses on cultivating skills for constructive and transparent peer review practices.
Recognizing the pivotal role peer review plays in determining which research is published, funded, and ultimately shared with the scientific community, HHMI and PREreview have developed training sessions that emphasize mitigating biases that are often unintentionally embedded in the review process. PREReview’s specialized workshops, like “Open Reviewers” for manuscript reviews and “Open Grant Reviewers” for grant application reviews, address issues of DEI. These sessions provide participants with critical frameworks to identify and reduce the impact of biases, challenging participants to rethink their assumptions and adopt a fairer evaluation approach.
In addition to training, PREreview provides a range of open access resources, available on Zenodo under a CC BY 4.0 license, to guide reviewers in their practice. These resources help reviewers hone their skills, train others, and foster collaborative efforts within research communities. This partnership between HHMI and PREreview is a proactive approach in equipping the next generation of scientists with the tools needed to conduct peer review that is not only rigorous but also equitable and unbiased.
ASAPbio is actively working to diversify and expand the peer review pool by fostering public engagement with preprint reviews through initiatives like the previously discussed Crowd Preprint Review and with the transformation of journal clubs into preprint review clubs. ASAPbio began experimenting with crowd review models in 2021. In this model, groups of researchers provided public feedback on preprints, initially focusing on cell biology topics, resulting in 14 reviews. The high engagement observed encouraged ASAPbio to continue and expand the initiative.
By 2022, ASAPbio coordinated 3 focused groups that provided public reviews for preprints in cell biology and biochemistry on bioRxiv, as well as infectious disease research in Portuguese on SciELO Preprints. This effort produced 27 reviews on bioRxiv and 13 on SciELO, illustrating the demand and potential impact of crowd-sourced peer review. In 2023, ASAPbio scaled up its efforts by organizing four crowds led by ASAPbio Fellows, which collectively generated 34 public reviews. The initiative’s success has inspired ASAPbio to continue crowd preprint review activities into 2024 with new long-term groups covering areas such as cell biology, immunology, microbiology, and meta-research.
In addition to crowd reviews, ASAPbio has launched a fund to support traditional journal clubs in becoming preprint review clubs, enabling early-career researchers who already discuss and evaluate research papers in group settings to formally share their reviews with authors and the wider community. By facilitating and financially supporting this transformation, ASAPbio aims to establish a proof-of-concept for integrating journal clubs into the preprint peer review ecosystem, potentially creating a sustainable model for broadening peer review participation and improving feedback mechanisms across research disciplines.
These initiatives are helping to reshape the peer review process by bringing more diverse voices into the conversation and providing opportunities for early-career researchers to develop their skills. By focusing on training, inclusivity, and transparency, these efforts are laying the groundwork for a better trained and more diverse peer review system.
Conclusion
The peer review process, long regarded as the backbone of scholarly integrity and quality, is undergoing a transformative shift as new platforms and initiatives like PREreview, ReviewCommons, PCI, Sciety, preLights, ASAPbio, and PubPeer introduce fresh ideas and experimental approaches. They are supported by an evolving technical ecosystem that incorporates open protocols and system-to-system communication methods that enhance research transferability and interoperability of peer review components. Leveraging technology that enables seamless communication between diverse systems and incorporating persistent identifiers like ORCID and DOIs, these models are built on modern, adaptable workflows that integrate smoothly into existing frameworks. These innovations extend beyond efficiency, striving to create a peer review system that values equity, collaboration, and transparency. Beyond structural improvements, these initiatives are dedicated to enriching the peer review ecosystem by training early-career researchers, engaging underrepresented groups, and fostering a broader, more diverse pool of reviewers, thereby bringing varied expertise and perspectives to research evaluation.
By embracing community-driven approaches, open standards, and inclusivity-focused training programs, the peer review process is becoming better suited to today’s fast-paced and interconnected research landscape. As illustrated by these use cases, decentralized peer review has the power to accelerate scientific progress, elevate research quality, and ensure a more inclusive and trustworthy evaluation process, ultimately positioning peer review to meet the demands of contemporary scholarship.
Acknowledgements
I would like to thank the following people for their help reviewing this article and providing feedback:
Denis Bourguet, Peer Community In (PCI); Monica Granados, PREreview; Fiona Hutton, eLife, Sciety; Katherine Brown, Company of Biologists, preLights; Richard Sever, bioRxiv, medRxiv and Cold Spring Harbor Laboratory; Emily Esten, Knowledge Futures, DocMaps; Mariia Levchenko, Europe PMC; Ben Mudrak, American Chemical Society, ChemRxiv.
References and Links
- https://www.niso.org/standards-committees/meca
- https://www.coar-repositories.org/what-we-do/notify/
- https://docmaps.knowledgefutures.org/
- https://www.crossref.org/
- https://datacite.org/
- https://doi.org/10.31222/osf.io/yu4sm
- https://doi.org/10.31222/osf.io/yu4sm
- https://prereview.org/
- https://www.biorxiv.org/
- https://eeb.embo.org/about
- https://blog.sciety.org/join-as-a-group-on-sciety/
Tony Alves (https://orcid.org/0000-0001-7054-1732) is SVP, Product Management, HighWire Press.
Opinions expressed are those of the authors and do not necessarily reflect the opinions or policies of their employers, the Council of Science Editors, or the Editorial Board of Science Editor.
